Conformational suppression of inter-receptor signaling defects by Peter Ames and Sandy Parkinson |
|
| Motile bacteria follow gradients of attractant and repellent chemicals with high sensitivity. The cells' gradient-sensing chemoreceptors are physically clustered and may function as a cooperative signaling array. Although native chemoreceptor molecules are typically transmembrane homodimers, the dimers appear to associate through their cytoplasmic tips to form trimers of dimers, which may be an important architectural element in the assembly and operation of receptor clusters. In a previous study (Ames et al., 2002), we found that certain mutations (designated Tsr*) at the trimer contact sites of the serine chemoreceptor not only abrogated Tsr function, but also blocked the function of heterologous receptors. To explore the mechanistic basis of these jamming effects, we isolated and characterized mutations (designated Tar^) in the aspartate chemoreceptor that restored function to Tsr* receptors. Some examples of the suppression effects between Tsr* and Tar^ receptors are shown in Figure 1. | |
Figure 1. Chemotaxis phenotypes of Tar^ and Tsr* mutants. Cells containing various combinations of Tsr and Tar receptors were tested for chemotactic behavior on tryptone soft agar plates. Colony morphologies are shown after incubation for 10 hours at 32.5°C. Top panel: Jamming behavior of Tsr* mutants. The doubly wild-type control colony (center) forms two cell bands or rings reflecting serine chemotaxis (Tsr function; outer ring) and aspartate chemotaxis (Tar function; inner ring). Bottom panels: Suppression of the epistatic Tsr* defects by Tar^ mutations. Tar^-1 suppresses only the Tsr*-1 and Tsr*-3 defects. Note that these colonies have no aspartate ring. Tar^-2 suppresses all but the Tsr*-1 defect. Note that these colonies exhibit two chemotactic rings. Tar^-2 evidently retains Tar function in the presence of suppressible Tsr* receptors. |
 |
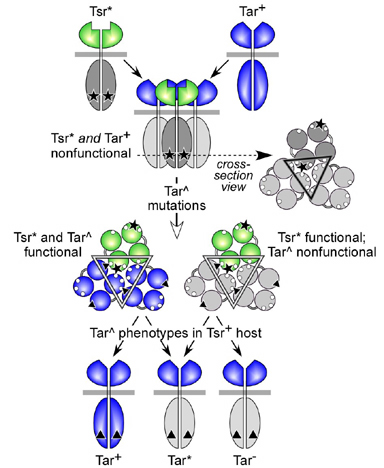
| A working model of Tar^-Tsr* suppression is presented in Figure 2. We propose that restoration of Tsr* function occurs through compensatory Tar^-induced conformational changes in mixed trimers of Tar^ and Tsr* dimers. To test this idea, we examined the pattern of Tar^- Tsr* suppression effects. A hallmark of conformational suppression mechanisms is their allele-specificity, reflecting the underlying structural specificity of protein-protein interactions. | |
Figure 2. Working model of inter-receptor jamming and suppression. Receptor dimers with signaling domains in color are functional; dimers with grayed signaling domains are not. According to the model, Tsr receptors with jamming lesions (Tsr*) block the function of wild-type Tar receptors by forming defective mixed trimers of dimers. The Tsr* amino acid replacement (black star) affects one of the trimer contact residues (small white circles). Tar^ mutations (black triangles) impart a compensatory conformational change to mixed trimers that restores the signaling ability of some Tsr* receptors. Some Tar^ mutants retain signaling function in the presence of their Tsr* partner, others do not. Some of the nonfunctional Tar^ receptors are themselves able to jam wild-type Tsr receptors. |
 |
| We found that Tar^- Tsr* suppression was indeed allele-specific (Fig. 3). Each Tsr* allele responded to a small group of Tar^ suppressors. Moreover, all Tar^ alterations occurred in or near the Tar trimer contact residues, the most direct and structurally specific way to perturb dimer-dimer interactions within trimers. | |
Figure 3. Allele-specificity of Tar^- Tsr* suppression. The 19 Tar^ mutations in this study fell into six suppression classes, each illustrated with a backbone view of the tip and trimer contact region of the Tsr/Tar signaling domain. Tsr* mutations are indicated by black, space-filled alpha carbons of the mutant residues. The residues altered in their Tar^ suppressors are shown with space-filled alpha carbons on the same dimer. Yellow residues denote Tar^ receptors with wild-type output signals; green residues denote CW-biased Tar^ mutants; red residues denote CCW-biased Tar^ mutants. |
 |
| These results provide strong support for the trimer-of-dimers model of collaborative receptor signaling teams, which are thought to be responsible for the high-gain signaling properties of bacterial chemoreceptors (see related posters). | |
| Ames, P. and J.S. Parkinson (2006) Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. USA 102: 9292-9297. | |
| Ames, P., C.A. Studdert, R.H. Reiser, and J.S. Parkinson (2002). Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99: 7060-7065. | |
